ASD Screening Classification Project
CLASSIFICATION OF AUTISTIC SPECTRUM DISORDER SCREENING DATA
Autism Spectrum Disorder (ASD) refers to a range of conditions characterized by challenges with social skills, repetitive behaviors, speech and non-verbal communication According to the Centers for Disease Control, autism affects an estimated 1 in 59 children in the United States. In 2013 the American Psychiatric Association merged four distinct diagnoses into one under the name of Autism Spectrum Disorder, there were previously referred to as Autistic Disorder, Childhood Disintegrate Disorder, Pervasive Development Disorder-Not Otherwise Specified (PDD-NOS) and Asperger Syndrome.
For a long time, ASD was associated with a lengthy process to get properly diagnosed. With an increase in the number of ASD cases around the world, a faster screening tool was created called the Autism Spectrum Quotient (AQ) consisting of 50 questions. A condensed version, the AQ-10, was created as a faster self-diagnosis tool to determine an individual’s position on the autism-normality spectrum. While the AQ-10 is NOT used for a definitive diagnosis, a score greater that 6/10 would be a flag that you should seek a professional diagnosis.
Objective
While the tool is a great first step, there aren’t many available datasets associated with clinical screenings and behavior. Per Fadi Fayez Thabtah, creator of the set, most available datasets on autism are genetic in nature. This set consists of the responses to behavior questions on the AQ-10 tool along with the results. In addition, 10 individual characteristics were made part of this set which have been used by the behavioral sciences experts for ASD detection. I intend to also use classification to determine the effectiveness of not just the AQ-10 tool but also the individual traits provided in diagnosing ASD by using classification and clustering algorithms. I will be using Decision Trees, Naïve Bayes, k-NN and Support Vector Machines.
Import Libraries & Data
Lets import the R packages we will be using
#----- Install Packages ----#
if (!require(C50)) install.packages('C50') #C5.0 Decision Trees and Rule-Based Models
if (!require(caret)) install.packages('caret') #Classification and Regression Training
if (!require(countrycode)) install.packages('countrycode') #Convert Country Names and Country Codes
if (!require(class)) install.packages('class') #Functions for Classification
if (!require(dplyr)) install.packages('dplyr') #for Data Manipulation
if (!require(e1071)) install.packages('e1071') #naive Bayes classifier
if (!require(farff)) install.packages('farff') #Reads and writes 'ARFF' files
if (!require(gmodels)) install.packages('gmodels') #for Model Fitting
if (!require(kernlab)) install.packages('kernlab') #Support Vector Machines
if (!require(sqldf)) install.packages('sqldf') #Manipulate R Data Frames Using SQL
Next I downloaded and imported the data. The data consists of three separate data sets available from the UCI Machine Learning Repository. The sets are divided by age; children (4-11), adolescents (12-17), and adult (18 and over). The number of observations for each of the sets were 292 (children), 104 (adolescents), and 704 (adults) combining for 1100 total observations. The attributes were: ASD Diagnosis (our classifier), age (in years), gender, ethnicity, born with jaundice (Y/N), Family member with ASD (Y/N), Relation of the person completing the test, Country of Residence, Use of a screening application previously, age description, Questions 1-10 of the AQ tool (each separate), and the Screening Score on the AQ-10. The set was distributed as an ARFF file.
The sets can be downloaded directly via the links below:
ASD Screening Children Data Set
ASD Screening Adolescent Data Set
ASD Screening Adult Data Set
Data Preparation
There was some heavy amount of preparation that I needed to performed before I could analyze the data. Because each of the three datasets were small (child, adolescent and adult) my first step was to merge the three data sets into one larger set. There were several NA values that existed in the age column as well as in some of the factor columns (for example ethnicity). For the age NAs, I used the avg age in each of the primary sets (child adolescent, adult) to replace the NA value before merging into one set. For factor columns I created an “Unknown Value”. Several columns also needed to be recategorized to numeric value in order to be able to use them in the various algorithms.
Once I had one dataset, I did some additional cleaning of the data. The age column was eliminated since there was already a categorical “age description” column. Since research shows that ASD is not specific to country or ethnicity, I decided to remove those columns as well. Country of origin specifically had 89 different discrete values for this column, so this made the data easier to work with. The remaining values were converted into discrete values, I did this by creating separate variables for each columns option and using 1 and 0 to indicate Yes or No. Had I kept the countries and ethnicities; this would have resulted in over 100 additional variables.
# Sets for editing
AutChild <- Autism_Child_Data
AutTeen <- Autism_Adolescent_Data
AutAdult <- Autism_Adult_Data
# Combining the three sets into one total set
AutTotal <- rbind(AutChild,AutTeen,AutAdult)
summary(AutTotal)
'data.frame': 1100 obs. of 21 variables:
Questionaire Data
Create Test and Training Sets
#----- Creating Training and Test Sets FOR ONLY Questionaire-----#
Total1 <- AutTotal[,c(1, 8:18)]
# Create sets using caret library
inTraining <- createDataPartition(Total1$ClassASD, times = 1, p = .66, list = FALSE)
trainAut <- Total1[ inTraining,]
trainAut$ClassASD <- droplevels( trainAut)$ClassASD
testAut <- Total1[-inTraining,]
testAut$ClassASD <- droplevels(testAut)$ClassASD
# Predicting factor is a score >6 (results)
Build Model using c50 Algorithmn (c50 library)
I created a Decision Tree using the c50 library’s C5.0 algorithm. I first attempted to create a decision tree based on just the survey and final score. My intention was to see if there were any patterns with the answers, however I was surprised with the results. The resulting decision tree was completely based on the result of the survey, <=6 was no expected diagnosis of autism and >6 was expecting a Yes. Upon using the test data on the model, the model accurately predicted the outcome diagnosis 100% of the time. This raised a red flag as we should never expect 100% accuracy. All other subsequent classification algorithms also produced a 100% Accuracy in prediction, this caused me to want to NOT look at the questionnaire responses and instead look at all the other variables.
# building Model using c50 Algorithim (c50 library)
dt_model <- C5.0(trainAut[-1], trainAut$ClassASD)
# evaluating performance
pred_model <- predict(dt_model, testAut)
CrossTable(testAut$ClassASD, pred_model, prop.chisq = FALSE, prop.c = FALSE, prop.r = FALSE, dnn = c('actual diagnosis', 'predicted diagnosis') )
Call:
C5.0.default(x = trainAut[-1], y = trainAut$ClassASD)
Classification Tree
Number of samples: 727
Number of predictors: 11
Tree size: 2
Non-standard options: attempt to group attributes
Call:
C5.0.default(x = trainAut[-1], y = trainAut$ClassASD)
C5.0 [Release 2.07 GPL Edition] Tue Apr 28 16:49:55 2020
-------------------------------
Class specified by attribute `outcome'
Read 727 cases (12 attributes) from undefined.data
Decision tree:
result <= 6: NO (467)
result > 6: YES (260)
Evaluation on training data (727 cases):
Decision Tree
----------------
Size Errors
2 0( 0.0%) <<
(a) (b) <-classified as
---- ----
467 (a): class NO
260 (b): class YES
Attribute usage:
100.00% result Time: 0.0 secs
Cell Contents
|-------------------------|
| N |
| N / Table Total |
|-------------------------|
Total Observations in Table: 373
| predicted diagnosis
actual diagnosis | NO | YES | Row Total |
-----------------|-----------|-----------|-----------|
NO | 240 | 0 | 240 |
| 0.643 | 0.000 | |
-----------------|-----------|-----------|-----------|
YES | 0 | 133 | 133 |
| 0.000 | 0.357 | |
-----------------|-----------|-----------|-----------|
Column Total | 240 | 133 | 373 |
-----------------|-----------|-----------|-----------|
All Variables Data (Excluding Questionaire Questions)
I decided to create a new set for testing using all the existing variables in the set, with the exception of the questionairre responses.
Create Data Set
In order to do this I had to expand the existing categorical data and convert it into binary data. This was a lengthy process and is covered in detail in the code at the end of this project. This end result was the creation of the set AutExp2 consisting of 35 columns 1100 rows.
Create Test and Training Sets
#----- Creating Traing and Test Sets FOR ALL FACTORS (No quetionaire)-----#
Total2 <- AutExp2[,c(1,19:35)]
# create sets using caret library
inTraining <- createDataPartition(Total2$ClassASD, times = 1, p = .66, list = FALSE)
trainAut <- Total2[ inTraining,]
trainAut$ClassASD <- droplevels( trainAut)$ClassASD
testAut <- Total2[-inTraining,]
testAut$ClassASD <- droplevels(testAut)$ClassASD
Build Model using c50 Algorithmn (c50 library)
I created a new model using all the other variables excluding the survey question and results. The decision tree model created had a probability of being accurate 67% of the time. Upon running my test set. The tree was able to accurately predict the diagnosis 68.63% (237/373) of the time. Below is the resulting decision tree model. As you can see, the attributes that contributed the most to this tree was the age description (12-17 age group) and the relationship of the person who filled out the form. It also seems that the algorithm is having problems correctly predicted “Yes ASD” outcomes. It correctly identified 80.83% of “No ASD” outcome but only correctly predicted 46.61% of “Yes ASD” outcome. Also, when attempting to boost, the number of trials stopped at 2 since the classifier was not accurate. There was no improvement in boosting. For this data, the decision tree is not a suitable tool for classification. There may possibly be additional variables not accounted for in this data that would aid in better classification via a Decision Tree.
# building Model using c50 Algorithim (c50 library)
dt_model <- C5.0(trainAut[-1], trainAut$ClassASD)
summary(dt_model)
# evaluating performance
pred_model <- predict(dt_model, testAut)
CrossTable(testAut$ClassASD, pred_model, prop.chisq = FALSE, prop.c = FALSE, prop.r = FALSE, dnn = c('actual diagnosis', 'predicted diagnosis') )
# improving model performance
dt_boost10 <- C5.0(trainAut[-1], trainAut$ClassASD, trials= 10)
dt_boost10 #use summary(dt_boost10) to get the % accuracy and tree
summary(dt_boost10)
# evaluating boosted performance
pred_model_boost <- predict(dt_boost10, testAut)
CrossTable(testAut$ClassASD, pred_model_boost, prop.chisq = FALSE, prop.c = FALSE, prop.r = FALSE, dnn = c('actual diagnosis', 'predicted diagnosis') )
Call:
C5.0.default(x = trainAut[-1], y = trainAut$ClassASD)
Classification Tree
Number of samples: 727
Number of predictors: 17
Tree size: 2
Non-standard options: attempt to group attributes
Call:
C5.0.default(x = trainAut[-1], y = trainAut$ClassASD)
C5.0 [Release 2.07 GPL Edition] Tue Apr 28 16:49:56 2020
-------------------------------
Class specified by attribute `outcome'
Read 727 cases (18 attributes) from undefined.data
Decision tree:
12to17 <= 0: NO (658/216)
12to17 > 0: YES (69/25)
Evaluation on training data (727 cases):
Decision Tree
----------------
Size Errors
2 241(33.1%) <<
(a) (b) <-classified as
---- ----
442 25 (a): class NO
216 44 (b): class YES
Attribute usage:
100.00% 12to17 Time: 0.0 secs
Cell Contents
|-------------------------|
| N |
| N / Table Total |
|-------------------------|
Total Observations in Table: 373
| predicted diagnosis
actual diagnosis | NO | YES | Row Total |
-----------------|-----------|-----------|-----------|
NO | 224 | 16 | 240 |
| 0.601 | 0.043 | |
-----------------|-----------|-----------|-----------|
YES | 114 | 19 | 133 |
| 0.306 | 0.051 | |
-----------------|-----------|-----------|-----------|
Column Total | 338 | 35 | 373 |
-----------------|-----------|-----------|-----------|
Call:
C5.0.default(x = trainAut[-1], y = trainAut$ClassASD, trials = 10)
Classification Tree
Number of samples: 727
Number of predictors: 17
Number of boosting iterations: 10
Average tree size: 2.6
Non-standard options: attempt to group attributes
Call:
C5.0.default(x = trainAut[-1], y = trainAut$ClassASD, trials = 10)
C5.0 [Release 2.07 GPL Edition] Tue Apr 28 16:49:56 2020
-------------------------------
Class specified by attribute `outcome'
Read 727 cases (18 attributes) from undefined.data
----- Trial 0: -----
Decision tree:
12to17 <= 0: NO (658/216)
12to17 > 0: YES (69/25)
----- Trial 1: -----
Decision tree:
RelationUnkown > 0: NO (99.8/22.6)
RelationUnkown <= 0:
:...18Plus <= 0: YES (241.2/103.6)
18Plus > 0: NO (386.1/149.2)
----- Trial 2: -----
Decision tree:
RelationUnkown > 0: NO (95.4/25.8)
RelationUnkown <= 0:
:...FamAutismN <= 0: YES (108.8/46.1)
FamAutismN > 0: NO (522.8/232)
----- Trial 3: -----
Decision tree:
RelationUnkown > 0: NO (92.6/27.9)
RelationUnkown <= 0:
:...JundiceN <= 0: YES (98.4/42.8)
JundiceN > 0:
:...f <= 0: NO (299.3/129.8)
f > 0: YES (236.7/109.9)
----- Trial 4: -----
Decision tree:
RelationParent <= 0: NO (509.8/221.6)
RelationParent > 0: YES (217.2/99.8)
----- Trial 5: -----
Decision tree:
RelationUnkown > 0: NO (88.4/30.9)
RelationUnkown <= 0:
:...12to17 <= 0: NO (570/275.7)
12to17 > 0: YES (68.6/29.5)
----- Trial 6: -----
Decision tree:
RelationUnkown <= 0: YES (653.7/315.5)
RelationUnkown > 0: NO (55.3)
----- Trial 7: -----
Decision tree:
RelationUnkown > 0: NO (52.6)
RelationUnkown <= 0:
:...JundiceN <= 0: YES (101.1/47)
JundiceN > 0: NO (554.4/267.3)
----- Trial 8: -----
Decision tree:
RelationUnkown > 0: NO (49.8)
RelationUnkown <= 0:
:...18Plus <= 0: YES (291.3/113.5)
18Plus > 0: NO (268.9/61.1)
----- Trial 9: -----
Decision tree:
NO (559/173.2)
Evaluation on training data (727 cases):
Trial Decision Tree
----- ----------------
Size Errors
0 2 241(33.1%)
1 3 246(33.8%)
2 3 260(35.8%)
3 4 297(40.9%)
4 2 260(35.8%)
5 3 238(32.7%)
6 2 398(54.7%)
7 3 260(35.8%)
8 3 246(33.8%)
9 1 260(35.8%)
boost 236(32.5%) <<
(a) (b) <-classified as
---- ----
425 42 (a): class NO
194 66 (b): class YES
Attribute usage:
100.00% RelationParent
100.00% RelationUnkown
100.00% 12to17
85.56% JundiceN
85.56% FamAutismN
85.56% 18Plus
73.45% f
Time: 0.0 secs
Cell Contents
|-------------------------|
| N |
| N / Table Total |
|-------------------------|
Total Observations in Table: 373
| predicted diagnosis
actual diagnosis | NO | YES | Row Total |
-----------------|-----------|-----------|-----------|
NO | 219 | 21 | 240 |
| 0.587 | 0.056 | |
-----------------|-----------|-----------|-----------|
YES | 104 | 29 | 133 |
| 0.279 | 0.078 | |
-----------------|-----------|-----------|-----------|
Column Total | 323 | 50 | 373 |
-----------------|-----------|-----------|-----------|
Boost Model
# improving model performance
dt_boost10 <- C5.0(trainAut[-1], trainAut$ClassASD, trials= 10)
dt_boost10 #use summary(dt_boost10) to get the % accuracy and tree
summary(dt_boost10)
# evaluating boosted performance
pred_model_boost <- predict(dt_boost10, testAut)
CrossTable(testAut$ClassASD, pred_model_boost, prop.chisq = FALSE, prop.c = FALSE, prop.r = FALSE, dnn = c('actual diagnosis', 'predicted diagnosis') )
Call:
C5.0.default(x = trainAut[-1], y = trainAut$ClassASD, trials = 10)
Classification Tree
Number of samples: 727
Number of predictors: 17
Number of boosting iterations: 10
Average tree size: 2.6
Non-standard options: attempt to group attributes
Call:
C5.0.default(x = trainAut[-1], y = trainAut$ClassASD, trials = 10)
C5.0 [Release 2.07 GPL Edition] Tue Apr 28 16:49:56 2020
-------------------------------
Class specified by attribute `outcome'
Read 727 cases (18 attributes) from undefined.data
----- Trial 0: -----
Decision tree:
12to17 <= 0: NO (658/216)
12to17 > 0: YES (69/25)
----- Trial 1: -----
Decision tree:
RelationUnkown > 0: NO (99.8/22.6)
RelationUnkown <= 0:
:...18Plus <= 0: YES (241.2/103.6)
18Plus > 0: NO (386.1/149.2)
----- Trial 2: -----
Decision tree:
RelationUnkown > 0: NO (95.4/25.8)
RelationUnkown <= 0:
:...FamAutismN <= 0: YES (108.8/46.1)
FamAutismN > 0: NO (522.8/232)
----- Trial 3: -----
Decision tree:
RelationUnkown > 0: NO (92.6/27.9)
RelationUnkown <= 0:
:...JundiceN <= 0: YES (98.4/42.8)
JundiceN > 0:
:...f <= 0: NO (299.3/129.8)
f > 0: YES (236.7/109.9)
----- Trial 4: -----
Decision tree:
RelationParent <= 0: NO (509.8/221.6)
RelationParent > 0: YES (217.2/99.8)
----- Trial 5: -----
Decision tree:
RelationUnkown > 0: NO (88.4/30.9)
RelationUnkown <= 0:
:...12to17 <= 0: NO (570/275.7)
12to17 > 0: YES (68.6/29.5)
----- Trial 6: -----
Decision tree:
RelationUnkown <= 0: YES (653.7/315.5)
RelationUnkown > 0: NO (55.3)
----- Trial 7: -----
Decision tree:
RelationUnkown > 0: NO (52.6)
RelationUnkown <= 0:
:...JundiceN <= 0: YES (101.1/47)
JundiceN > 0: NO (554.4/267.3)
----- Trial 8: -----
Decision tree:
RelationUnkown > 0: NO (49.8)
RelationUnkown <= 0:
:...18Plus <= 0: YES (291.3/113.5)
18Plus > 0: NO (268.9/61.1)
----- Trial 9: -----
Decision tree:
NO (559/173.2)
Evaluation on training data (727 cases):
Trial Decision Tree
----- ----------------
Size Errors
0 2 241(33.1%)
1 3 246(33.8%)
2 3 260(35.8%)
3 4 297(40.9%)
4 2 260(35.8%)
5 3 238(32.7%)
6 2 398(54.7%)
7 3 260(35.8%)
8 3 246(33.8%)
9 1 260(35.8%)
boost 236(32.5%) <<
(a) (b) <-classified as
---- ----
425 42 (a): class NO
194 66 (b): class YES
Attribute usage:
100.00% RelationParent
100.00% RelationUnkown
100.00% 12to17
85.56% JundiceN
85.56% FamAutismN
85.56% 18Plus
73.45% f
Time: 0.0 secs
Cell Contents
|-------------------------|
| N |
| N / Table Total |
|-------------------------|
Total Observations in Table: 373
| predicted diagnosis
actual diagnosis | NO | YES | Row Total |
-----------------|-----------|-----------|-----------|
NO | 219 | 21 | 240 |
| 0.587 | 0.056 | |
-----------------|-----------|-----------|-----------|
YES | 104 | 29 | 133 |
| 0.279 | 0.078 | |
-----------------|-----------|-----------|-----------|
Column Total | 323 | 50 | 373 |
-----------------|-----------|-----------|-----------|
Build Naive Bayes Model
Next, I applied the Naïve Bayes algorithm via the e1071 package. The Naïve Bayes performed better than the Decision Tree, I ran improved model with a laplace value of 3. The Naïve Bayes model performed significantly better than the decision tree. Since the classifier is not looking for interdependence, it was able to do a better job of classifying the data.
#----- Use Na?ve Bayes Model-----#
# build the classifiers
Autclassifier<- naiveBayes(trainAut, trainAut$ClassASD)
Autclassifier
# evaluate model classfier
Auttestpredicter <- predict(Autclassifier, testAut)
# compare predictions to true values
CrossTable(testAut$ClassASD, Auttestpredicter, prop.chisq = FALSE, prop.t = FALSE, dnn = c('actual diagnosis', 'predicted diagnosis') )
# create confusion matrix
cMatrix <- table(Auttestpredicter, testAut$ClassASD)
plot(cMatrix)
confusionMatrix(cMatrix)
Naive Bayes Classifier for Discrete Predictors
Call:
naiveBayes.default(x = trainAut, y = trainAut$ClassASD)
A-priori probabilities:
trainAut$ClassASD
NO YES
0.6423659 0.3576341
Conditional probabilities:
ClassASD
trainAut$ClassASD NO YES
NO 1 0
YES 0 1
f
trainAut$ClassASD [,1] [,2]
NO 0.4304069 0.4956641
YES 0.4692308 0.5000148
m
trainAut$ClassASD [,1] [,2]
NO 0.5695931 0.4956641
YES 0.5307692 0.5000148
JundiceN
trainAut$ClassASD [,1] [,2]
NO 0.8715203 0.3349820
YES 0.8076923 0.3948736
JundiceY
trainAut$ClassASD [,1] [,2]
NO 0.1284797 0.3349820
YES 0.1923077 0.3948736
FamAutismN
trainAut$ClassASD [,1] [,2]
NO 0.8822270 0.3226848
YES 0.8038462 0.3978521
FamAutismY
trainAut$ClassASD [,1] [,2]
NO 0.1177730 0.3226848
YES 0.1961538 0.3978521
RelationHealthcarePro
trainAut$ClassASD [,1] [,2]
NO 0.01070664 0.1030278
YES 0.01923077 0.1376000
RelationOther
trainAut$ClassASD [,1] [,2]
NO 0.004282655 0.06537174
YES 0.007692308 0.08753632
RelationParent
trainAut$ClassASD [,1] [,2]
NO 0.2141328 0.4106593
YES 0.3846154 0.4874425
RelationRelative
trainAut$ClassASD [,1] [,2]
NO 0.04710921 0.2120996
YES 0.05384615 0.2261492
RelationSelf
trainAut$ClassASD [,1] [,2]
NO 0.5374732 0.4991285
YES 0.4653846 0.4997623
RelationUnkown
trainAut$ClassASD [,1] [,2]
NO 0.18629550 0.3897625
YES 0.06923077 0.2543357
UsedAppN
trainAut$ClassASD [,1] [,2]
NO 0.9785867 0.1449128
YES 0.9769231 0.1504374
UsedAppY
trainAut$ClassASD [,1] [,2]
NO 0.02141328 0.1449128
YES 0.02307692 0.1504374
12to17
trainAut$ClassASD [,1] [,2]
NO 0.05353319 0.2253356
YES 0.16923077 0.3756788
18Plus
trainAut$ClassASD [,1] [,2]
NO 0.7194861 0.4497321
YES 0.4807692 0.5005936
4to11
trainAut$ClassASD [,1] [,2]
NO 0.2269807 0.4193292
YES 0.3500000 0.4778895
Cell Contents
|-------------------------|
| N |
| N / Row Total |
| N / Col Total |
|-------------------------|
Total Observations in Table: 373
| predicted diagnosis
actual diagnosis | NO | YES | Row Total |
-----------------|-----------|-----------|-----------|
NO | 228 | 12 | 240 |
| 0.950 | 0.050 | 0.643 |
| 0.991 | 0.084 | |
-----------------|-----------|-----------|-----------|
YES | 2 | 131 | 133 |
| 0.015 | 0.985 | 0.357 |
| 0.009 | 0.916 | |
-----------------|-----------|-----------|-----------|
Column Total | 230 | 143 | 373 |
| 0.617 | 0.383 | |
-----------------|-----------|-----------|-----------|
Confusion Matrix and Statistics
Auttestpredicter NO YES
NO 228 2
YES 12 131
Accuracy : 0.9625
95% CI : (0.9378, 0.9793)
No Information Rate : 0.6434
P-Value [Acc > NIR] : < 2e-16
Kappa : 0.9196
Mcnemar's Test P-Value : 0.01616
Sensitivity : 0.9500
Specificity : 0.9850
Pos Pred Value : 0.9913
Neg Pred Value : 0.9161
Prevalence : 0.6434
Detection Rate : 0.6113
Detection Prevalence : 0.6166
Balanced Accuracy : 0.9675
'Positive' Class : NO
Improve Model
# build improved model
Autclassifier2<- naiveBayes(trainAut, trainAut$ClassASD, laplace = 3)
Autclassifier2
# evaluate improved model
Auttestpredicter2 <- predict(Autclassifier2, testAut)
# compare improved predictions to true values
CrossTable(testAut$ClassASD, Auttestpredicter2, prop.chisq = FALSE, prop.t = FALSE, dnn = c('actual diagnosis', 'predicted diagnosis') )
Naive Bayes Classifier for Discrete Predictors
Call:
naiveBayes.default(x = trainAut, y = trainAut$ClassASD, laplace = 3)
A-priori probabilities:
trainAut$ClassASD
NO YES
0.6423659 0.3576341
Conditional probabilities:
ClassASD
trainAut$ClassASD NO YES
NO 0.993657505 0.006342495
YES 0.011278195 0.988721805
f
trainAut$ClassASD [,1] [,2]
NO 0.4304069 0.4956641
YES 0.4692308 0.5000148
m
trainAut$ClassASD [,1] [,2]
NO 0.5695931 0.4956641
YES 0.5307692 0.5000148
JundiceN
trainAut$ClassASD [,1] [,2]
NO 0.8715203 0.3349820
YES 0.8076923 0.3948736
JundiceY
trainAut$ClassASD [,1] [,2]
NO 0.1284797 0.3349820
YES 0.1923077 0.3948736
FamAutismN
trainAut$ClassASD [,1] [,2]
NO 0.8822270 0.3226848
YES 0.8038462 0.3978521
FamAutismY
trainAut$ClassASD [,1] [,2]
NO 0.1177730 0.3226848
YES 0.1961538 0.3978521
RelationHealthcarePro
trainAut$ClassASD [,1] [,2]
NO 0.01070664 0.1030278
YES 0.01923077 0.1376000
RelationOther
trainAut$ClassASD [,1] [,2]
NO 0.004282655 0.06537174
YES 0.007692308 0.08753632
RelationParent
trainAut$ClassASD [,1] [,2]
NO 0.2141328 0.4106593
YES 0.3846154 0.4874425
RelationRelative
trainAut$ClassASD [,1] [,2]
NO 0.04710921 0.2120996
YES 0.05384615 0.2261492
RelationSelf
trainAut$ClassASD [,1] [,2]
NO 0.5374732 0.4991285
YES 0.4653846 0.4997623
RelationUnkown
trainAut$ClassASD [,1] [,2]
NO 0.18629550 0.3897625
YES 0.06923077 0.2543357
UsedAppN
trainAut$ClassASD [,1] [,2]
NO 0.9785867 0.1449128
YES 0.9769231 0.1504374
UsedAppY
trainAut$ClassASD [,1] [,2]
NO 0.02141328 0.1449128
YES 0.02307692 0.1504374
12to17
trainAut$ClassASD [,1] [,2]
NO 0.05353319 0.2253356
YES 0.16923077 0.3756788
18Plus
trainAut$ClassASD [,1] [,2]
NO 0.7194861 0.4497321
YES 0.4807692 0.5005936
4to11
trainAut$ClassASD [,1] [,2]
NO 0.2269807 0.4193292
YES 0.3500000 0.4778895
Cell Contents
|-------------------------|
| N |
| N / Row Total |
| N / Col Total |
|-------------------------|
Total Observations in Table: 373
| predicted diagnosis
actual diagnosis | NO | YES | Row Total |
-----------------|-----------|-----------|-----------|
NO | 223 | 17 | 240 |
| 0.929 | 0.071 | 0.643 |
| 0.978 | 0.117 | |
-----------------|-----------|-----------|-----------|
YES | 5 | 128 | 133 |
| 0.038 | 0.962 | 0.357 |
| 0.022 | 0.883 | |
-----------------|-----------|-----------|-----------|
Column Total | 228 | 145 | 373 |
| 0.611 | 0.389 | |
-----------------|-----------|-----------|-----------|
Build k-NN Model
Next, I used the class package to apply the K-NN classification algorithm. The original unscaled version of the model I created only produced a 62.4% accuracy when tested. Like the Decision Tree Model, it has an issue with over reporting the “No-ASD” classification. I attempted to improve the model by rescaling using z-score standardization. This improved the accuracy slightly to 68.63%. The issue is still with the misclassification of the “Yes-ASD” outcome.
#----- Use k-NN Model-----#
# model training
trainAutlabel <- trainAut[,1]
testAutlabel <- testAut [,1]
knnAutSpread <- knn(train=trainAut[,-1], testAut [,-1], cl=trainAutlabel, k=7)
summary(knnAutSpread)
CrossTable(x=testAutlabel, y=knnAutSpread, prop.chisq = FALSE)
Cell Contents
|-------------------------|
| N |
| N / Row Total |
| N / Col Total |
| N / Table Total |
|-------------------------|
Total Observations in Table: 373
| knnAutSpread
testAutlabel | NO | YES | Row Total |
-------------|-----------|-----------|-----------|
NO | 206 | 34 | 240 |
| 0.858 | 0.142 | 0.643 |
| 0.675 | 0.500 | |
| 0.552 | 0.091 | |
-------------|-----------|-----------|-----------|
YES | 99 | 34 | 133 |
| 0.744 | 0.256 | 0.357 |
| 0.325 | 0.500 | |
| 0.265 | 0.091 | |
-------------|-----------|-----------|-----------|
Column Total | 305 | 68 | 373 |
| 0.818 | 0.182 | |
-------------|-----------|-----------|-----------|
Improve Performance
# improved performance using z-score standardization
# new model training
Total2z <- as.data.frame(scale(Total2[,-1]))
Total2z_train <- Total2z[1:733,]
Total2z_test <- Total2z[734:1100,]
Total2z_train_label <- Total2[1:733,1]
Total2z_test_label <- Total2[734:1100,1]
# new performance
knnAutPrediction <- knn(train=Total2z_train, Total2z_test, cl=Total2z_train_label, k=11)
summary(knnAutPrediction)
CrossTable(x=Total2z_test_label, y=knnAutPrediction, prop.chisq = FALSE)
Cell Contents
|-------------------------|
| N |
| N / Row Total |
| N / Col Total |
| N / Table Total |
|-------------------------|
Total Observations in Table: 367
| knnAutPrediction
Total2z_test_label | NO | YES | Row Total |
-------------------|-----------|-----------|-----------|
NO | 247 | 26 | 273 |
| 0.905 | 0.095 | 0.744 |
| 0.744 | 0.743 | |
| 0.673 | 0.071 | |
-------------------|-----------|-----------|-----------|
YES | 85 | 9 | 94 |
| 0.904 | 0.096 | 0.256 |
| 0.256 | 0.257 | |
| 0.232 | 0.025 | |
-------------------|-----------|-----------|-----------|
Column Total | 332 | 35 | 367 |
| 0.905 | 0.095 | |
-------------------|-----------|-----------|-----------|
Build SVM Model
Lastly, I attempted classify using the kernlab package to use the SVM algorithm. When running the model using the vanilladot (basic linear) kernel, the model had a 65.68% accuracy in predicting the outcome. I attempted to improve the model’s performance by using the rbfdot (Radial basis/Gaussian), however this resulted in less accuracy (64.87%)
#----- Use SVM Model-----#
# build the model classifier
ksvmmodelclassifier <- ksvm(ClassASD ~ ., data=trainAut, kernel ="vanilladot")
ksvmmodelclassifier
# evaluate performance
ksmvmodelpredictions <- predict(ksvmmodelclassifier, testAut)
CrossTable(testAut$ClassASD, ksmvmodelpredictions, prop.chisq = FALSE, prop.c = FALSE, prop.r = FALSE, dnn = c('actual number', 'predicted number') )
# model predicted 236 / 373 (63.27%)
Setting default kernel parameters
Support Vector Machine object of class "ksvm"
SV type: C-svc (classification)
parameter : cost C = 1
Linear (vanilla) kernel function.
Number of Support Vectors : 505
Objective Function Value : -482.1433
Training error : 0.331499
Cell Contents
|-------------------------|
| N |
| N / Table Total |
|-------------------------|
Total Observations in Table: 373
| predicted number
actual number | NO | YES | Row Total |
--------------|-----------|-----------|-----------|
NO | 224 | 16 | 240 |
| 0.601 | 0.043 | |
--------------|-----------|-----------|-----------|
YES | 114 | 19 | 133 |
| 0.306 | 0.051 | |
--------------|-----------|-----------|-----------|
Column Total | 338 | 35 | 373 |
--------------|-----------|-----------|-----------|
Improve Model
# improved performance
ksvmmodelclassifierrbf <- ksvm(ClassASD ~ ., data=trainAut, kernel ="rbfdot")
ksvmmodelclassifierrbf
ksmvmodelpredictionsrbf <- predict(ksvmmodelclassifierrbf, testAut)
CrossTable(testAut$ClassASD, ksmvmodelpredictionsrbf, prop.chisq = FALSE, prop.c = FALSE, prop.r = FALSE, dnn = c('actual number', 'predicted number') )
# model predicted 247/ 373 (66.21%)
Support Vector Machine object of class "ksvm"
SV type: C-svc (classification)
parameter : cost C = 1
Gaussian Radial Basis kernel function.
Hyperparameter : sigma = 0.0490338963559971
Number of Support Vectors : 506
Objective Function Value : -466.0458
Training error : 0.290234
Cell Contents
|-------------------------|
| N |
| N / Table Total |
|-------------------------|
Total Observations in Table: 373
| predicted number
actual number | NO | YES | Row Total |
--------------|-----------|-----------|-----------|
NO | 210 | 30 | 240 |
| 0.563 | 0.080 | |
--------------|-----------|-----------|-----------|
YES | 97 | 36 | 133 |
| 0.260 | 0.097 | |
--------------|-----------|-----------|-----------|
Column Total | 307 | 66 | 373 |
--------------|-----------|-----------|-----------|
Results
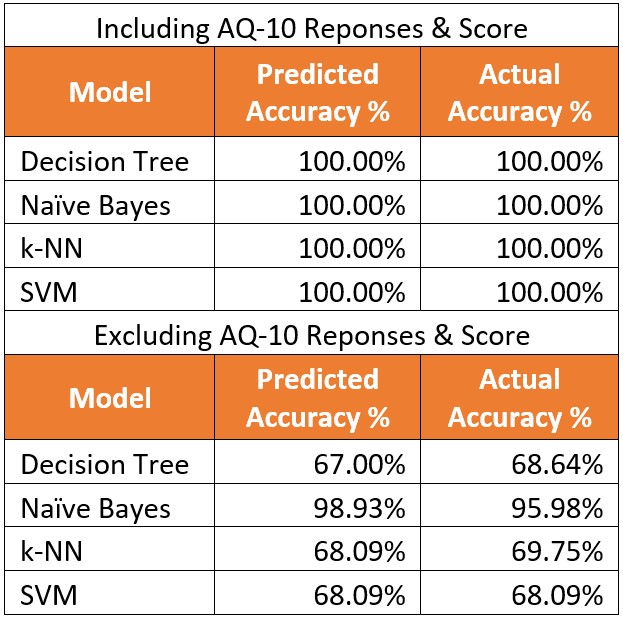
I found the results very odd. It seems that when taking into account the AQ-10 responses and score, all the algorithms were able to classify. ASD outcome 100% of the time. When looking at all the behavioral factors, there was a steep decline in the accuracy, with the exception of Naïve Bayes.
Conclusion
So how is this possible? Is the AQ-10 really that good of a predicter? Are there more factors at play than the variables in this data set? I looked up a few research articles, to see what information was available regarding the AQ tool. A study in the Journal of Autism and Development Disorders concluded that there is little difference in results of the AQ-10 and AQ-50 and the AQ-10 can be a potentially useful screening tool (Booth, etal 2013). A later study in Psychological Medicine concluded that not were AQ scores not a significant predictor of diagnosis for ASD, but that the 64% of those who scored below the cut off and had a “No-ASD” outcome were actually false negatives who were later in fact diagnosed with ASD (Ashwood, etal, 2016). This led me to look at the provided data sets more. Upon further inspection, I noticed that the data was bias. All individuals with an AQ score of >=6 later received a No diagnosis. There were no signs of any false positives or negatives. This explains why there was a 100% accurate prediction when using using the AQ-10 responses and score.
With regards to the other factors, I believe there are two issues at play. First there needs to be more data as there isn’t a variance in results. Second, since ASD is still being studied I believe that there are several other variables at play which aren’t included in this data set, and may prove as better predictors to classify ASD. I believe more data should be collected, including expanding the variables being recorded.
References
Ashwood, K. L., Gillan, N., Horder, J., Hayward, H., Woodhouse, E., McEwen, F. S., . . . Murphy, D. G. (2016). Predicting the diagnosis of autism in adults using the autism-spectrum quotient (AQ) questionnaire. Psychological Medicine, 46(12), 2595-2604. doi:10.1017/S0033291716001082
Booth, T., Murray, A. L., McKenzie, K., Kuenssberg, R., O’Donnell, M., & Burnett, H. (2013). Brief report: An evaluation of the AQ-10 as a brief screening instrument for ASD in adults. Journal of Autism and Developmental Disorders, 43(12), 2997-3000. doi:10.1007/s10803-013-1844-5
Code
You can check out the full R code using the following methods:
- Github Page: Francisco’s Repository
- Google Colab: